(1) Submitting and evaluating results
To upload and evaluate the result of your assembly, genome or taxon binning, or taxonomic profile for a CAMI dataset against its underlying genomes or gold standards, please follow these instructions.
Simplified instructions for the impatient
- Format your file for the specific task category: assembly, binning, or profiling
- Go to the Upload and evaluate page from the Submit menu and drag and drop the file onto the designated area, or click the area to open a system dialog to select the file. Wait until the upload completes. Upon successful upload, you will be redirected automatically to another page.
- On the next page, select the target dataset and samples if the file contains an assembly. If it is a binning or profiling file, the target dataset samples will be selected automatically from the SampleID identifiers.
- Select whether the evaluation results should be public (default), meaning that they will be added to the respective results page (menu Results) and visible to all users upon completion of the evaluation. In this case, please fill out the metadata form, allowing for reproduction of the results.
- Lastly, hit the Evaluate button. The evaluation will start immediately or be queued, and its status will be displayed on the same page. It typically takes a few seconds to complete for binning and profiling and up to a few hours for assembly. Refresh the page from time to time to check for the completion.
In more detail
First, format your file for the specific task category. For assembly, use FASTA; for binning and profiling, use one of the tab-separated formats described here (binning) and here (profiling). Make sure the binning and profiling file contains the SampleID identifier(s) of the target dataset sample(s). Binning and profiling results of multiple samples of the same dataset should be included in a single file, by concatenation. Zip-compression, with one file per zip, is supported for all task categories, tar.gz is not!
Go to the Upload and evaluate page using the website menu, as shown in the picture. If you wish to make your results public and visible for other users online, or not have them deleted after a few weeks, you can optionally log in first.
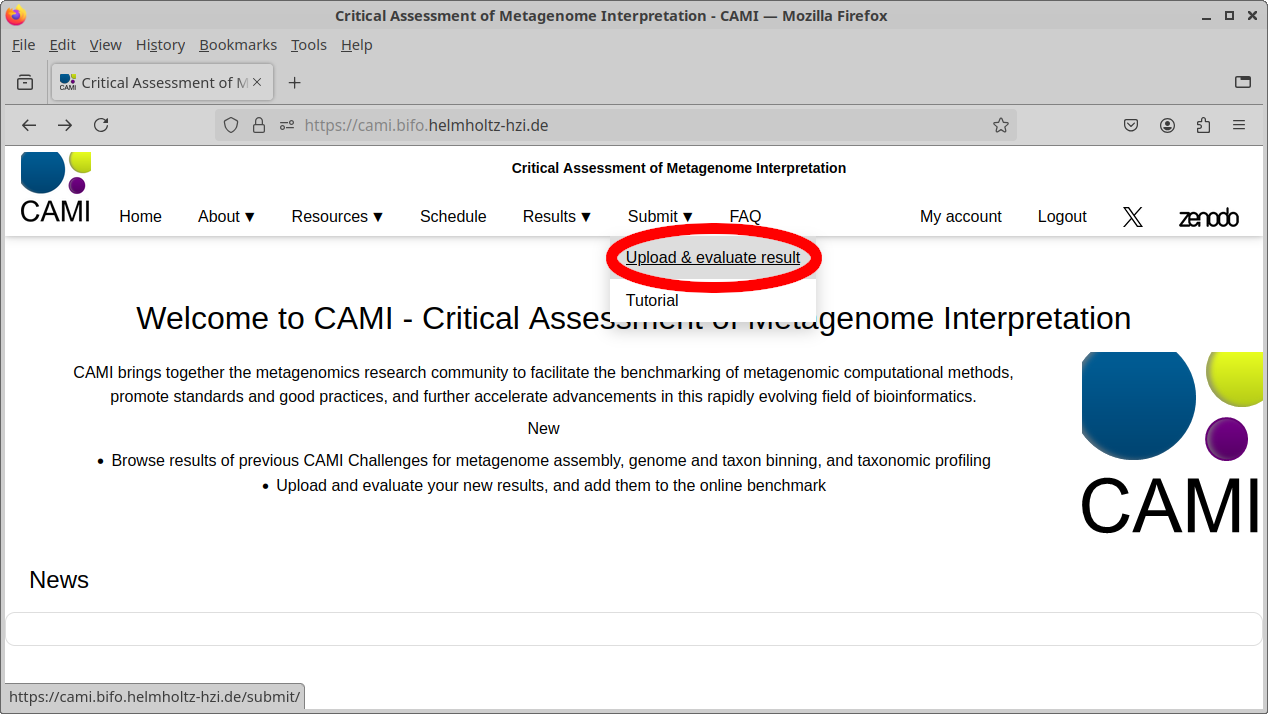
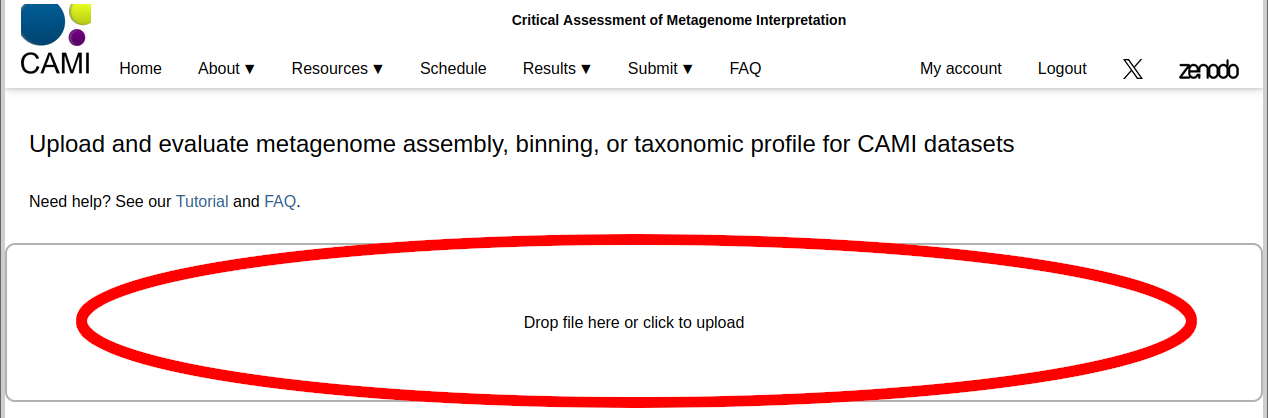
On the Upload and evaluate page, drag and drop the file onto the area shown below, or click the area to open a system dialog to select the file. Wait until the upload completes. Upon successful upload, you will be redirected automatically to another page. The upload will fail if the file is formatted incorrectly or contains invalid SampleID identifiers. If the problem persists, please contact us.
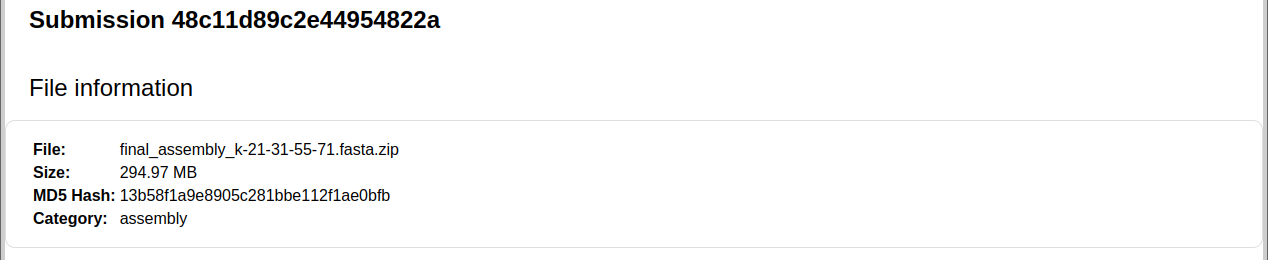
Depending on the file size, the next page may show a message stating that the file is being processed. Please wait a few seconds to a couple of minutes and refresh the page. Once this completes, the submission will be identified by a unique code and basic file information will be shown, as in the picture below. You can verify if the file was uploaded correctly by checking its file size and MD5 Hash code. On a Linux system, you can generate an MD5 Hash code for a file using the command:
md5sum <filename>
The hash code is unique for the file and should match the code displayed on the page, for your upload. Example:
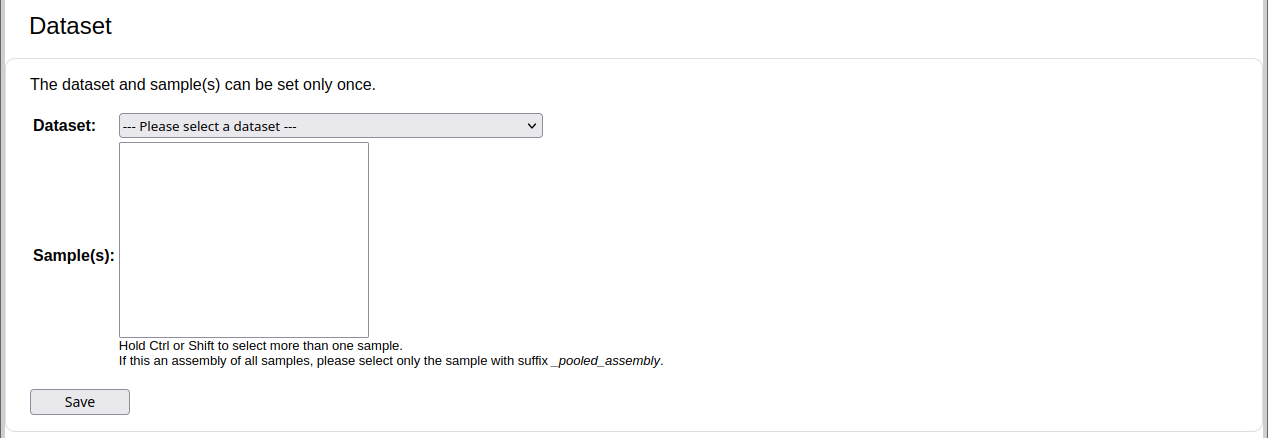
Next, select the target dataset and sample(s) if the file contains an assembly. For co-assemblies of multiple samples, multiple samples can be selected by holding down Ctrl while clicking on the sample names or by using the Shift key. The assembly will be evaluated using MetaQUAST against the underlying genomes of the selection. Optionally, click on the Save button, or just continue with the next step. Note that this can only be set once. If you select the wrong dataset and samples, you will need to re-upload the file as another submission.
If it is a binning or profiling file, the target dataset samples will be selected automatically from the SampleID identifiers. In this case, it will be evaluated using AMBER and OPAL, respectively. The figure shows an example allowing for the selection of the target dataset and sample, for an assembly.
Now, select in the metadata form whether the evaluation results should be public (default), meaning that they will be added to the respective results page (menu Results) and visible to all users upon completion of the evaluation. In this case, please fill out the rest of the metadata form, allowing for reproduction of the results. Note that, in any case, your file will *not* be available for download by other users. To share it, you can provide a download link from a third-party repository in the comments and DOI fields, in the metadata form.
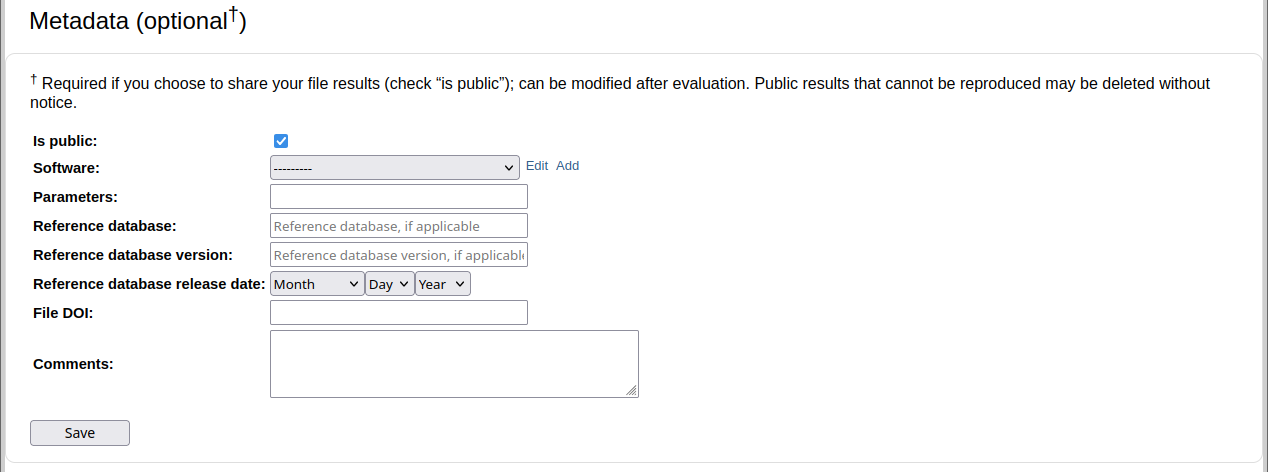
Lastly, the page allows to select evaluation parameters for MetaQUAST, AMBER, or OPAL, depending on the submission type. We recommend to use default parameters.
For assembly: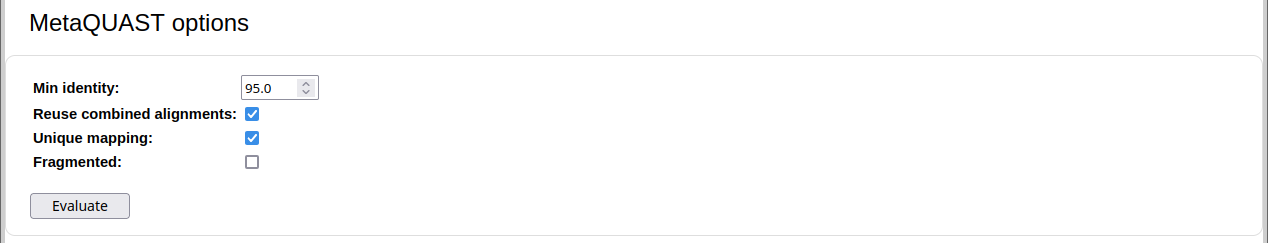 For genome and taxon binning:
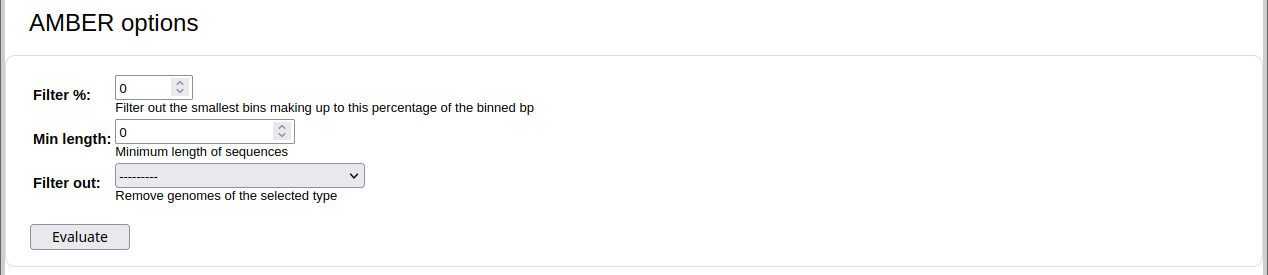
For genome and taxon binning:
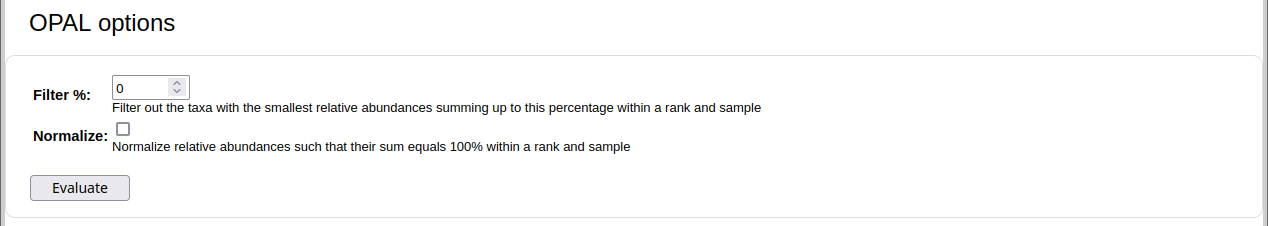
 For profiling:
For profiling:
Hit the Evaluate button. The evaluation will start immediately or be queued, and its status will be displayed on the same page. It typically takes a few seconds to complete for binning and profiling and up to a few hours for assembly. Refresh the page from time to time to check for the completion. It is possible to repeat the evaluation with different parameters once the current evaluation completes.
(2) Interpreting results
Public results are ranked within their category (assembly, taxon or genome binning, or taxonomic profiling), sample, and taxonomic rank (if applicable). They are accessible through the Results option in the navigation menu on the page header above. Results per metric are compared with each other to produce a heatmap-like table, with blue indicating better performance and red worse. Gray indicates metric values around the median. Example:
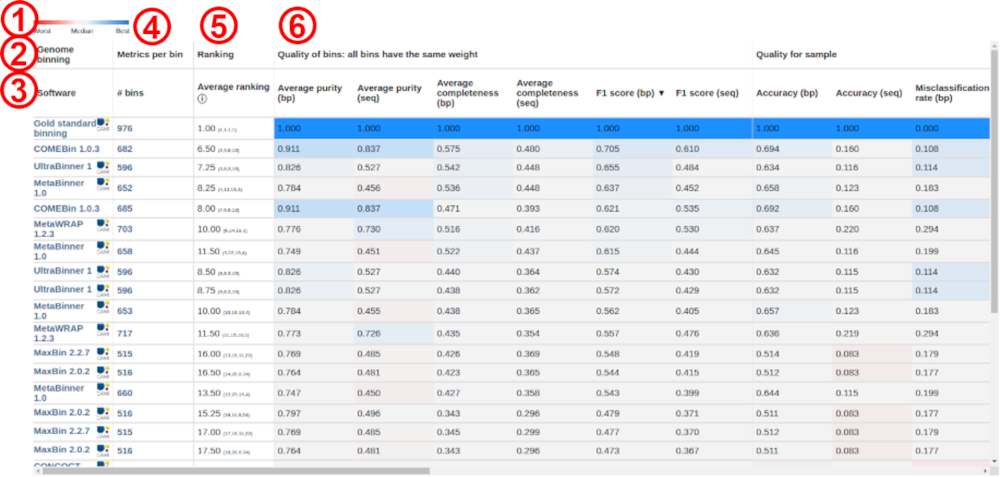
In this example above, the numbers indicate the following:
- Performance heatmap color scale
- Category (here, genome binning)
- Evaluated software and version. Clicking on the linked software takes the user to the page of the respective result submission and exact evaluation, where software parameters and other metadata are available, as provided by the submitter. A result submission can be evaluated several times by the submitter with different MetaQUAST, AMBER, or OPAL parameters, depending on the category. For this reason, the same software can appear several times on the table. A different submission for the same software is also possible.
- Metrics per bin (in the case of binning). Shown are the numbers of bins predicted by the respective software. They are clickable and take the user to a page with detailed results per bin. In the case of taxonomic profiling, detailed results per taxon are available.
- Performance rankings. These are the averages of the ranking (small numbers beside the average) per several metrics. Move the mouse pointer over the information symbol to see which metrics are considered in the ranking. The best results with a metric ranks 1, the 2nd best, 2, and so on.
- Metrics groups and metrics. For metrics definitions, we refer the user to the publications of MetaQUAST, AMBER, and OPAL, as well as of the CAMI Challenges.
The tables layouts are identical for all software categories, but the metrics are category specific. In all cases, the tables are sortable by all metrics and performance ranks, by clicking on the respective metric.

